a book whose cover i can judge …
January 30th, 2009 by Lawrence David
i don’t think i’ve blogged about this topic yet; with the aid of some wonderful experimental collaborators and my advisor, i co-authored a paper in science this past summer on microbial ecology. my contribution to the project was an algorithm i named AdaptML. it’s a pretty cool algorithm*: i think it’s one of the first to gracefully combine genetic sequence information with the metadata describing which environments those data were sampled from, in order to ultimately produce a model of what environments bacteria hosting those sequences are adapted to living in. the algorithm can even predict environmental associations for the ancestral bacteria which gave rise to the present day ones you’re studying.
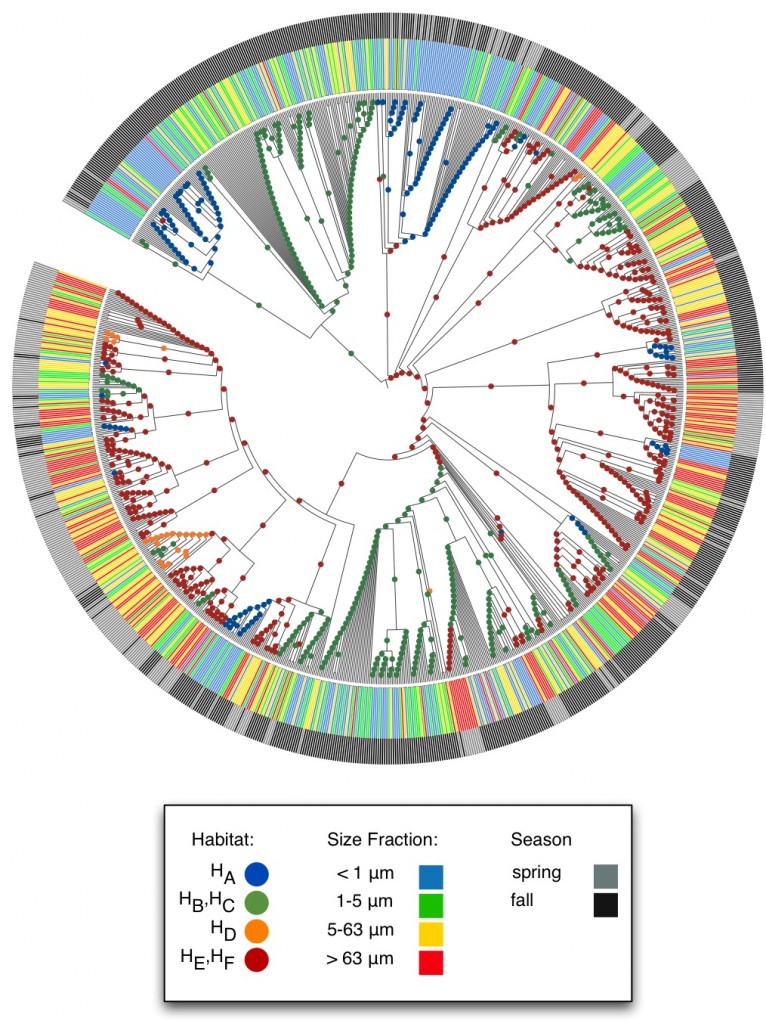
using a really nifty piece of online software called iToL**, i was able to plot the output of AdaptML in a very pleasing way. for instance, below is a sample figure used to help produce the science manuscript. the tree structure represents our best guess as to how a particular genus of bacteria named Vibrio evolved over hundreds of millions of years. (if Vibrio sounds familiar to you, it’s because one strain causes cholera.) the colors on the outer rings relate what kind of environment present day samples were found in; the inner balls tell you were we predict ancestral bacteria would have wanted to live.
in any case, the reason i bring all of this up (besides to feel good about myself for getting a paper published in a good journal), is because i learned today that the above figure made it onto the cover of a very well known textbook in genetics. it lives on amazon, and will now forever provide proof of my programming ineptitude if some bugs in my code eventually emerge.
* for those comp bio geeks who might be reading this, what’s nifty is what AdaptML does under the hood. the idea is that it treats the environmental metadata like sequence information and runs a general markov model to infer ancestral environmental metadata. for instance, if you had present day genetic sequences all related by a phylogenetic tree, you could apply a jukes-cantor or kimura model to calculate the most likely ancestral sequences on your tree, by assuming that each nucleotide in your sequence has a certain frequency and that the probabilities of nucleotides transitions satisfy certain properties. AdaptML uses that same idea, except that instead of considering 4 possible character states (nucleotides), it can handle N states, where N is the number of observed environments.
** i can’t rave about this software enough. i can honestly say it has changed the way i interpret my data; i never would have noticed patterns of evolution embedded in my datasets, had i not looked at them via iToL.


I did not understand a single sentence in this post. Congrats on the neat cover photo. Reminds me of the apple spinning beach ball of death.
http://itsthevideo.com/ghrf/8y8y3ydhofn.htmlãƒãƒãƒ¼ã‚ティiphone 6s plusケース